Hypothesis Leanring Domain Growth#
\(_{Yongtao}\) \(_{Liu,}\)
\(_{youngtaoliu@gmail.com}\)
\(_{Feb}\) \(_{2024}\)
Install and Import#
import os
import win32com.client
import numpy as np
import time
import h5py
import sidpy
import pyNSID
import matplotlib.pyplot as plt
from tqdm import tqdm
# import acquition.py
from Acquisition_v0_5 import Acquisition # include the Acquistion_v0.py in the same directory
import cv2
# import imutils
from os.path import exists
# from jupyterthemes import jtplot
# jtplot.style()
Start BEPyAE.exe and set VI#
Start BEPyAE.ext
Set VI of BEPyAE; if this version includes PyScanner, also set VIs for PyScanner
newexp = Acquisition(exe_path = r"C:\BEPyAE 060123 01\BEPyAE.exe") # exe_path is the directory of BEPyAE;
Initialize Igor AR18#
Set offline development
Build a connection between BEPyAE and AR18
Get parameters in AR18
newexp.init_BEPyAE(offline_development = True) # set offline_development=True if doing offline development
# executing this will also initlize AR18
Set tip parameters#
set setpoint, tip locations
newexp.tip_control(tip_parms_dict = {"set_point_V_00": 1, "next_x_pos_00": -0.5, "next_y_pos_01": 0.5},
do_move_tip = True,
do_set_setpoint = True) # Executing this code will set setpoint to 1 V,
# and move tip to location [0.5, 0.5]
Setpoint is: 1.0
Tip parameters are: (-0.5, 0.5, 1.0)
Please reset if some parameters are incorrect
Set IO#
This defines IO parameters, such as AFM platform: AR18, amplifiers, channel data types, etc
newexp.define_io_cluster(IO_cluster_parms_dict = {"analog_output_amplifier_06": 1,
"channel_01_type_07": 1,
"channel_02_type_08": 2,"channel_03_type_09": 3,})
IO control parameters are: ('0 Cypher AR18', '6124', 4000000.0, 10.0, 10.0, 'AC and DC on AO0', 1.0, 'none', 'none', 'none', 'external')
Please reset if some parameters are incorrect
Set BE pulse parameters#
# set BE parameters
newexp.define_be_parms(be_parms_dict = {"center_frequency_Hz_00": 335, "band_width_Hz_01": 100,
"amplitude_V_02": 1, "phase_variation_03": 1,
"repeats_04": 4, "req_pulse_duration_s_05": 4,
"auto_smooth_ring_06": 1},
do_create_be_waveform = True)
BE parameters are: (335000.0, 100000.0, 1.0, 1.0, 4, 0.004, 1, 3352.2952763920002, 0.12159459061880915)
Please reset if some parameters are incorrect
(335000.0,
100000.0,
1.0,
1.0,
4,
0.004,
1,
3352.2952763920002,
0.12159459061880915)
BE Line scan to test BE parameters#
This is a single BE line scan
This returns 5 datasets: quick_fitting, complex spectra, and 3 channels
# Do a single line scan
qk_fit, com_spec, chn1, chn2, chn3 = newexp.do_line_scan(line_scan_parms_dict = {"num_BE_pulses_01": 32,
"start_x_pos_00": -0.5, "start_y_pos_01": 0,
"stop_x_pos_02": 0.5, "stop_y_pos_03": 0},
upload_to_daq = True, do_line_scan = True)
voltage offset and number of BE pulse are: (0.0, 32)
line scan start and end positions: (-0.5, 0.0, 0.5, 0.0)
Experiment Starts#
In this experiment, we begin by applying a DC pulse to switch the ferroelectric polarization. Subsequently, a BEPFM (Bias-Enhanced Piezoresponse Force Microscopy) measurement is conducted to image the domain structure.#
To initiate the measurement process, we first need to determine the location for each individual measurement. For each measurement, a new location is chosen, requiring a location array to record all the measurements as demonstrated below.
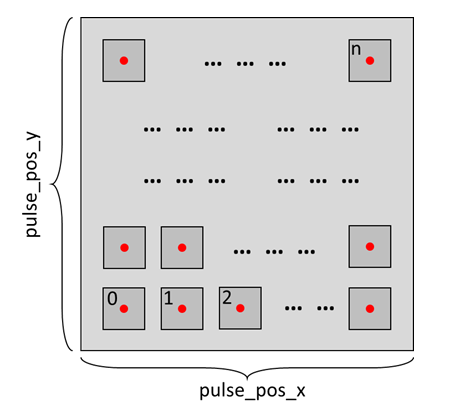
Run on local PC
Run below code on microscope computer.
Prior to expeirment, set a directory to save data#
os.chdir(r"C:\Hypothesis_learning\Test")
def domain_size (img, thresh):
thresh_img = np.copy(img)
thresh_img [img > thresh] = 1
thresh_img [img < thresh] = 0
# find contours in the thresholded image
thresh_img = thresh_img.astype(np.uint8)
cnts = cv2.findContours(thresh_img, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
cnts = imutils.grab_contours(cnts)
center = np.zeros((len(cnts), 2))
radius = np.zeros(len(cnts))
for num_domain in range (len(cnts)):
(x,y),r = cv2.minEnclosingCircle(cnts[num_domain])
#center location
center[num_domain,0] = x
center[num_domain,1] = y
#radius of minimum circle
radius[num_domain] = r
#calculate the distance between domain center and image center (writing point)
img_size = img.shape[0]
img_center_x = int(img_size/2)
img_center_y = int(img_size/2)
#away_writing_point = np.sqrt((center[:,0]-img_center_x)**2 + (center[:,1]-img_center_y)**2)
away_writing_point = np.sqrt((center[:,0]-32)**2 + (center[:,1]-32)**2)
#then, calculate the total area of residue domains
if len(radius) == 0:
print ("domain size is 0.0")
f, ax = plt.subplots()
ax.imshow(thresh_img)
plt.show()
plt.close()
return 0.0 #if no domains, return domain size as 0
elif away_writing_point.min() > 20: #if the nearest domain is away (13) from the center, treat it as noise or the domains originally in the region
print ("domain size is 0.0")
f, ax = plt.subplots()
ax.imshow(thresh_img)
plt.show()
plt.close()
return 0.0 #return domain size as 0
else:
#return_domain_size = np.sqrt((radius[away_writing_point.argmin()]**2)) # we use square root as area, treat the neareast domain as the written domain
return_domain_size = (radius.max())
f, ax = plt.subplots()
ax.imshow(thresh_img, origin = "lower")
# for c in range(len(radius)):
# Drawing_colored_circle=plt.Circle(center[c], radius[c], fill = False, color = 'white', linewidth = 4);
# ax.add_artist(Drawing_colored_circle)
Drawing_colored_circle=plt.Circle(center[away_writing_point.argmin()], radius[away_writing_point.argmin()],
fill = False, color = 'white', linewidth = 4);
ax.add_artist(Drawing_colored_circle)
plt.show()
plt.close()
return return_domain_size
Step 1. Generate a location array#
# All locations span across [start_point_x, end_point_x] in x-direction and [start_point_y, end_point_y] in y-direction.
# There are num_x rows and num_y columns in the locations array
start_point_x = -0.9 # Define location array parameters
end_point_x = 0.9
start_point_y = -0.9
end_point_y = 0.9
num_x = 10
num_y = 10
# Generate location array
pos_x = np.linspace(-0.9, 0.9, num_x)
pos_y = np.linspace(-0.9, 0.9, num_y)
pulse_pos = np.meshgrid(pos_x, pos_y)
pulse_pos_x = pulse_pos[0].reshape(-1)
pulse_pos_y = pulse_pos[1].reshape(-1) # pulse_pos_x and pulse_pos_y are the coordinates of all locations
# Set BEPFM image size
img_size = 0.1
# Check
if img_size > np.abs(pos_x[0]-pos_x[1]):
print ("Alert: there will be image overlap along x-direction")
elif img_size > np.abs(pos_y[0]-pos_y[1]):
print ("Alert: there will be image overlap along y-direction")
else:
print("{} locations are ready for experiments".format(len(pulse_pos_x)))
100 locations are ready for experiments
Run on GPU server
Run below code on GPU server.
Step 2. Install and import @ GPU server, and define help functions#
Please run this code @ GPU server
-------------------------------------------------------------------
!pip install --upgrade jax==0.2.25
!pip install -q git+https://github.com/ziatdinovmax/gpax.git
!pip install numpy --upgrade
from typing import Dict
import gpax
import numpyro
import numpy as onp
import jax.numpy as jnp
import jax.random as jra
import matplotlib.pyplot as plt
import time
from mlsocket import MLSocket
print('jax device: ', jax.devices())
gpax.utils.enable_x64()
File "C:\Users\yla\AppData\Local\Temp/ipykernel_21664/3515848405.py", line 3
-------------------------------------------------------------------
^
SyntaxError: invalid syntax
Please run this code @ GPU server
-------------------------------------------------------------------
def model_data(x: jnp.ndarray, params: Dict[str, float]) -> jnp.ndarray:
"""
r = r_c + r_0 * ((V/V_c)^2 - 1)^{1/3}
"""
return params["r_c"] + params["r_0"] * jnp.cbrt((x[:, 0] / params["V_c"])**2 - 1)
def grid2xy(X1, X2):
"""
Maps (M, N) grid to (M*N, 2) xy coordinates.
Removes NaNs (if any)
"""
X = jnp.concatenate((X1[None], X2[None]), 0)
d0, d1 = X.shape[0], X.shape[1] * X.shape[2]
X = X.reshape(d0, d1).T
X = X[~jnp.isnan(X).any(axis=1)]
return X
#data initialization and update functions
def init_training_data_exp(X, Y, num_seed_points=2, rng_seed=42, list_of_indices=None):
onp.random.seed(rng_seed)
indices = jnp.arange(len(X))
idx = list_of_indices
if idx is not None:
idx = onp.array(idx)
else:
idx = onp.random.randint(0, len(X), num_seed_points)
#idx = onp.unique(idx)
X_train, y_train = X[idx], Y
indices_train = indices[idx]
X_test = jnp.delete(X, idx, axis = 0)
#y_test = jnp.delete(Y, idx)
indices_test = jnp.delete(indices, idx)
return X_train, y_train, X_test, indices_train, indices_test
def update_datapoints_exp(next_point_idx, train, test, y_new):
"""Update "measured" dummy data points"""
X_train, y_train, indices_train = train
X_test, indices_test = test
X_train = jnp.append(X_train, X_test[next_point_idx][None], axis = 0)
X_test = jnp.delete(X_test, next_point_idx, axis = 0)
y_train = jnp.append(y_train, y_new[0])
#y_test = jnp.delete(y_test, next_point_idx)
indices_train = jnp.append(indices_train, next_point_idx)
indices_test = jnp.delete(indices_test, next_point_idx)
return (X_train, y_train, indices_train), (X_test, indices_test)
params = {"r_c": 1, "r_0": 1.5 , "V_c": 2}
#params = {"alpha": 1.1, "beta": .4}
d1 = 20
d2 = 20
V = jnp.linspace(1, 10, d1)
log_tau = jnp.linspace(-3, 2, d2)
V, log_tau = onp.meshgrid(V, log_tau)
X = grid2xy(V, log_tau)
y = model_data(X, params) + .4 * jra.normal(jra.PRNGKey(1), shape=(len(X),))
print ("Parameter space: ", X)
Step 3. Generate random seedings#
Please run this code @ GPU server
-------------------------------------------------------------------
# Generate seed write parameters
onp.random.seed(5)
seed_step = 20
# random index
idx = []
onp.random.seed(10)
idx = onp.random.choice(len(y), size = seed_step, replace=False)
print(len(set(idx)))
idx = onp.asarray(idx)
indx = jnp.asarray(idx).tolist()
print(len(idx))
X_measured = X[idx]
y_measured = y[idx]
X_unmeasured = jnp.delete(X, idx, axis=0)
onp.save("seed_write_paras.npy", X_measured) # send this to local
Run on local PC
Run below code on local PC.
Step 4. Transferseeding parameters from GPU server to local PC via sftp#
Step 5. Load seed writing parameters#
# load seed parameters from GPU-server
seed_paras_file = "/content/seed_write_paras.npy"
seed_write_paras = np.load(seed_paras_file)
# check seed parameters
print ("Seed Parameters: ", seed_write_paras)
Step 6. Start seed measurements#
seed_domain_size = []
for i in tqdm(range(len(seed_write_paras))):
#####################----------- Move tip to the pulse location -----------#####################
newexp.tip_control(tip_parms_dict = {"set_point_V_00": 1,
"next_x_pos_00": pulse_pos_x[i],
"next_y_pos_01": pulse_pos_y[i]},
do_move_tip = True, do_set_setpoint = True)
time.sleep(0.2)
#####################----------- Apply pulse -----------#####################
# load pulse
V_amp = -seed_write_paras[0][i]
V_time_log = seed_write_paras[1][i]
V_time = math.pow(1o, V_time)
print ("Write Parameters: {} V, {} s".format(V_amp, V_time))
# apply pulse
newexp.define_apply_pulse(pulse_parms_dict = {"pulse_init_amplitude_V_00": 0, "pulse_mid_amplitude_V_01": V_amp,
"pulse_final_amplitude_V_02": 0, "pulse_on_duration_s_03": V_time,
"rise_time_s_05": 1E-4, "pulse_final_duration_s_04": 20E-3,
"pulse_repeats_06": 1},
do_create_pulse = True, do_upload_pulse = True, do_apply_pulse = False)
#
time.sleep(1)
newexp.define_apply_pulse(pulse_parms_dict = {"pulse_init_amplitude_V_00": 0, "pulse_mid_amplitude_V_01": V_amp,
"pulse_final_amplitude_V_02": 0, "pulse_on_duration_s_03": V_time,
"rise_time_s_05": 1E-4, "pulse_final_duration_s_04": 20E-3,
"pulse_repeats_06": 1},
do_create_pulse = True, do_upload_pulse = True, do_apply_pulse = True)
time.sleep(2)
#####################----------- Do BEPFM to image domain -----------#####################
dset_pfm, dset_chns, dset_cs = newexp.raster_scan(raster_parms_dict = {"scan_pixel": 64,
"scan_x_start": pulse_pos_x[i]-(img_size/2),
"scan_y_start": pulse_pos_y[i]-(img_size/2),
"scan_x_stop": pulse_pos_x[i]+(img_size/2),
"scan_y_stop": pulse_pos_y[i]+(img_size/2)},
file_name = "HypoAl_Domain_Writing_{}".format(i),
progress_on = False, plot_on = False)
time.sleep(0.5)
# Plot BEPFM images
f, (ax1, ax2, ax3, ax4, ax5, ax6) = plt.subplots(1, 6, figsize = (30, 5), dpi = 100)
ax1.imshow(dset_pfm[:,:,0])
ax2.imshow(dset_pfm[:,:,1])
ax3.imshow(dset_pfm[:,:,2])
ax4.imshow(dset_pfm[:,:,3])
ax5.imshow(dset_chns[0,:,:])
ax6.imshow(dset_chns[1,:,:])
plt.show()
# Calculate domain size
pha = np.asarray(dset_pfm[:,:,3])
ds = domain_size (pha, 0.3)
seed_domain_size.append(ds) # add domain size to list
# Save seed domain size
np.save("seed_domain_size.npy", np.asarray(seed_domain_size)) # send this to GPU server
Run on GPU server
Run below code on GPU server.
Step 7. Send seeding results to GPU server via sftp#
Step 8. Prepare seeding train data#
Please run this code @ GPU server
-------------------------------------------------------------------
y_measured = onp.load("seed_domain_size.npy")
(X_train, y_train, X_test,
indices_train, indices_test) = init_training_data_exp(X, y_measured, list_of_indices = indx)
onp.savez("dataset_seed.npz", X_measured=X_train, y_measured=y_train,
indices_measured=indices_train, X_unmeasured=X_test, indices_unmeasured=indices_test)
onp.savez("dataset.npz", X_measured=X_train, y_measured=y_train,
indices_measured=indices_train, X_unmeasured=X_test, indices_unmeasured=indices_test)
Step 9. Start Hypothesis Active Learning exploration#
Please run this code @ GPU server
-------------------------------------------------------------------
exploration_step = 100
HOST = ''
PORT = 3446
# Bind to local PC
with MLSocket() as s:
s.bind((HOST, PORT))
s.listen()
conn, address = s.accept()
print("Bind successfully")
with conn:
# Warm-up phase
print('Warm-up starts')
# HypoAl Starts
for i in range(exploration_steps):
%run sGP_AL_domains_v1b.py dataset.npz
next_point_idx = onp.load("next_idx.npy")
nextpoint = onp.asarray(indices_test[next_point_idx])
print ("Next measurement location is: ", nextpoint)
# Send next point to local PC
time.sleep(0.01)
conn.send(nextpoint)
time.sleep(0.01)
print ("Waiting for new experiment result")
# Accept new experiment results
written_domain_size = conn.recv(920)
time.sleep(0.01)
# Update measured and unmeasured dataset
y_new = jnp.asarray([written_domain_size])
((X_train, y_train, indices_train), (X_test, indices_test)) = update_datapoints_exp(
next_point_idx, (X_train, y_train, indices_train), (X_test, indices_test), y_new)
# Save results
onp.savez("dataset{}.npz".format(i), X_measured=X_train, y_measured=y_train,
indices_measured=indices_train, X_unmeasured=X_test, indices_unmeasured=indices_test)
onp.savez("dataset.npz", X_measured=X_train, y_measured=y_train,
indices_measured=indices_train, X_unmeasured=X_test, indices_unmeasured=indices_test)
data = onp.load('dataset.npz')
print (data['X_measured'], '\n', data['y_measured'], '\n', data['indices_measured'])
print (data['X_unmeasured'], '\n', data['indices_unmeasured'])
# disconnet and shut down socket
conn.close()
s.shutdown(1)
s.close()
Run on local PC
Run below code on microscope computer.
Step 10. Connect to GPU server and start hypothesis driven active learning measurements#
exploration_step = 100
exploration_domain_size = []
HOST = 'localhost'
PORT = 9000
with MLSocket() as s:
s.connect((HOST, PORT))
for i in tqdm(range(exploration_step)):
#####################----------- Move tip to the pulse location -----------#####################
measure_loc = len (seed_write_paras) + i
newexp.tip_control(tip_parms_dict = {"set_point_V_00": 1,
"next_x_pos_00": pulse_pos_x[measure_loc],
"next_y_pos_01": pulse_pos_y[measure_loc]},
do_move_tip = True, do_set_setpoint = True)
time.sleep(0.2)
#####################----------- Apply pulse -----------#####################
# receive the first write parameters from Sockets
write_paras = s.recv(920)
V_amp = write_paras[0]
V_time = math.pow(10, write_paras[1])
# apply pulse
newexp.define_apply_pulse(pulse_parms_dict = {"pulse_init_amplitude_V_00": 0, "pulse_mid_amplitude_V_01": V_amp,
"pulse_final_amplitude_V_02": 0, "pulse_on_duration_s_03": V_time,
"rise_time_s_05": 1E-4, "pulse_final_duration_s_04": 20E-3,
"pulse_repeats_06": 1},
do_create_pulse = True, do_upload_pulse = True, do_apply_pulse = False)
time.sleep(1)
newexp.define_apply_pulse(pulse_parms_dict = {"pulse_init_amplitude_V_00": 0, "pulse_mid_amplitude_V_01": V_amp,
"pulse_final_amplitude_V_02": 0, "pulse_on_duration_s_03": V_time,
"rise_time_s_05": 1E-4, "pulse_final_duration_s_04": 20E-3,
"pulse_repeats_06": 1},
do_create_pulse = True, do_upload_pulse = True, do_apply_pulse = True)
time.sleep(1)
#####################----------- Do BEPFM to image domain -----------#####################
dset_pfm, dset_chns, dset_cs = newexp.raster_scan(raster_parms_dict = {"scan_pixel": 64,
"scan_x_start": pulse_pos_x[measure_loc]-(img_size/2),
"scan_y_start": pulse_pos_y[measure_loc]-(img_size/2),
"scan_x_stop": pulse_pos_x[measure_loc]+(img_size/2),
"scan_y_stop": pulse_pos_y[measure_loc]+(img_size/2)},
file_name = "Domain_Writing_{}".format(i),
progress_on = False, plot_on = False)
time.sleep(0.5)
# Plot BEPFM images
f, (ax1, ax2, ax3, ax4, ax5, ax6) = plt.subplots(1, 6, figsize = (30, 5), dpi = 100)
ax1.imshow(dset_pfm[:,:,0])
ax2.imshow(dset_pfm[:,:,1])
ax3.imshow(dset_pfm[:,:,2])
ax4.imshow(dset_pfm[:,:,3])
ax5.imshow(dset_chns[0,:,:])
ax6.imshow(dset_chns[1,:,:])
plt.show()
# Calculate domain size
pha = np.asarray(dset_pfm[:,:,3])
ds = domain_size (pha, 0.3)
# send domain size to GPU server
ds = np.asarray(ds)
time.sleep(0.01)
s.send(measured_point)
time.sleep(0.01)
exploration_domain_size.append(ds) # add domain size to list
# Save seed domain size
np.save("exploration_domain_size.npy", np.asarray(exploration_domain_size))
s.close()
Step 11. Do a BEPFM at the whole experiment area#
dset_pfm, dset_chns, dset_cs = newexp.raster_scan(raster_parms_dict = {"scan_pixel": 256, "scan_x_start": -1.0,
"scan_y_start": -1.0,"scan_x_stop": 1.0,
"scan_y_stop": 1.0}, file_name = "pfm_whole")
f, (ax1, ax2, ax3, ax4, ax5, ax6) = plt.subplots(1, 6, figsize = (30, 5), dpi = 100)
ax1.imshow(dset_pfm[:,:,0])
ax2.imshow(dset_pfm[:,:,1])
ax3.imshow(dset_pfm[:,:,2])
ax4.imshow(dset_pfm[:,:,3])
ax5.imshow(dset_chns[0,:,:])
ax6.imshow(dset_chns[1,:,:])
plt.show()
